Project
Information
Background
Among the living organisms, hyperthermophiles
belonging to the third domain of life, Archaea, are attractive
subjects of research from various aspects. Many efforts to understand
the strategies for adaptation to extremely high temperature
environments have revealed numerous physiologically intriguing and
unique properties of hyperthermophilic archaea. They have also
attracted attention from an industrial point of view as potential
resources for highly thermostable enzymes.
The euryarchaeal order Thermococcales, composed of
two major genera Thermococcus
and Pyrococcus may be the
best-studied hyperthermophiles. They are strictly anaerobic obligate
heterotrophs growing on complex proteinaceous substrates, and their
growth is strongly associated with the reduction of elemental sulfur.
Alternatively, with a few exceptions, they are capable of gaining
energy by fermentation of peptides, amino acids and sugars, forming
acids, CO2, and H2
in the absence of elemental sulfur.
The genus Pyrococcus,
with a higher optimum growth temperature (95-103 C) than Thermococcus (75-93 C), has
fascinated many microbiological researchers and has often been used
as the source organism for both fundamental and application-based
aspects of research. Therefore, although within the same genus, the
complete genome analyses of three species, P.
horikoshii (BA000001),
P. furiosus (AE009950),
and P. abyssi (AL096836),
have been performed. In contrast to Pyrococcus,
the genus Thermococcus
contains the highest number of characterized isolates. Recent
culture-dependent and culture-independent studies have indicated that
the members of Thermococcus
are more ubiquitously present in various deep-sea hydrothermal vent
systems than those of Pyrococcus. Consequently, Thermococcus strains, with their
larger population, are considered to play a major role in the ecology
and metabolic activity of microbial consortia within marine hot-water
ecosystems. However, despite the importance of this genus, no
complete genome sequence has been determined for Thermococcus. The Thermococcus genome can be
expected to encode genes responsible for various cellular functions
that provide an advantage for these strains in natural high
temperature habitats.
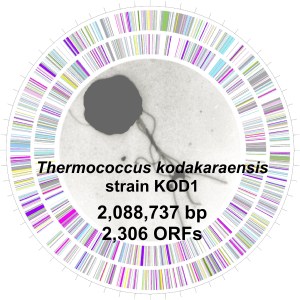
Features of
Thermococcus kodakaraensis strain KOD1
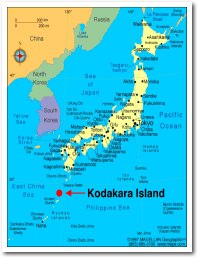
- Isolated from a solfatara on the shore of Kodakara Island,
Kagoshima, Japan as Pyrococcus
sp. strain KOD1 (Morikawa
et al. 1994)
- Re-classified as Thermococcus
kodakaraensis strain KOD1 based on phylogenetic and DNA-DNA
hybridization analyses (Atomi
et al. 2004)
- Taxonomoic
description
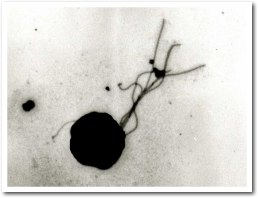
- Irregular cocci of 1-2µm diameter with several
flagella.
- Growth conditions: Temp. 60-100 C (opt. 85 C); pH 5-9 (opt.
6.5); Salt conc. 1-5% (opt. 3%); strict anaerobic
- Growth properties: Obligate heterotrophic
|
Carbon and energy sources
|
Sugar metabolism
|
Electron acceptor
|
Evolved gas
|
|
Amino acids, peptides
|
Gluconeogenesis
|
S0
|
H2S
|
|
Pyruvate
|
Gluconeogenesis
|
H+
|
H2
|
|
Starch
|
Glycolysis
|
H+
|
H2
|
- An abundant number of genes and their protein products from
this archaeon have been examined (Imanaka
and Atomi 2002), such as DNA polymerase (Hashimoto
et al. 2001), commercially available as an excellent enzyme
for PCR amplification from Toyobo
(in Japan), Novagen,
and Invitrogen.
- Useful gene disruption system, the first for a
hyperthermophile, has been developed (Sato
et al. 2003).
|